 BSc, MSc or PhD Thesis Projects - 2022
BSc, MSc or PhD Thesis Projects - 2022
Oferta de pràcticums, TFG, TFM o Tesis Doctoral a la Facultat de Biologia
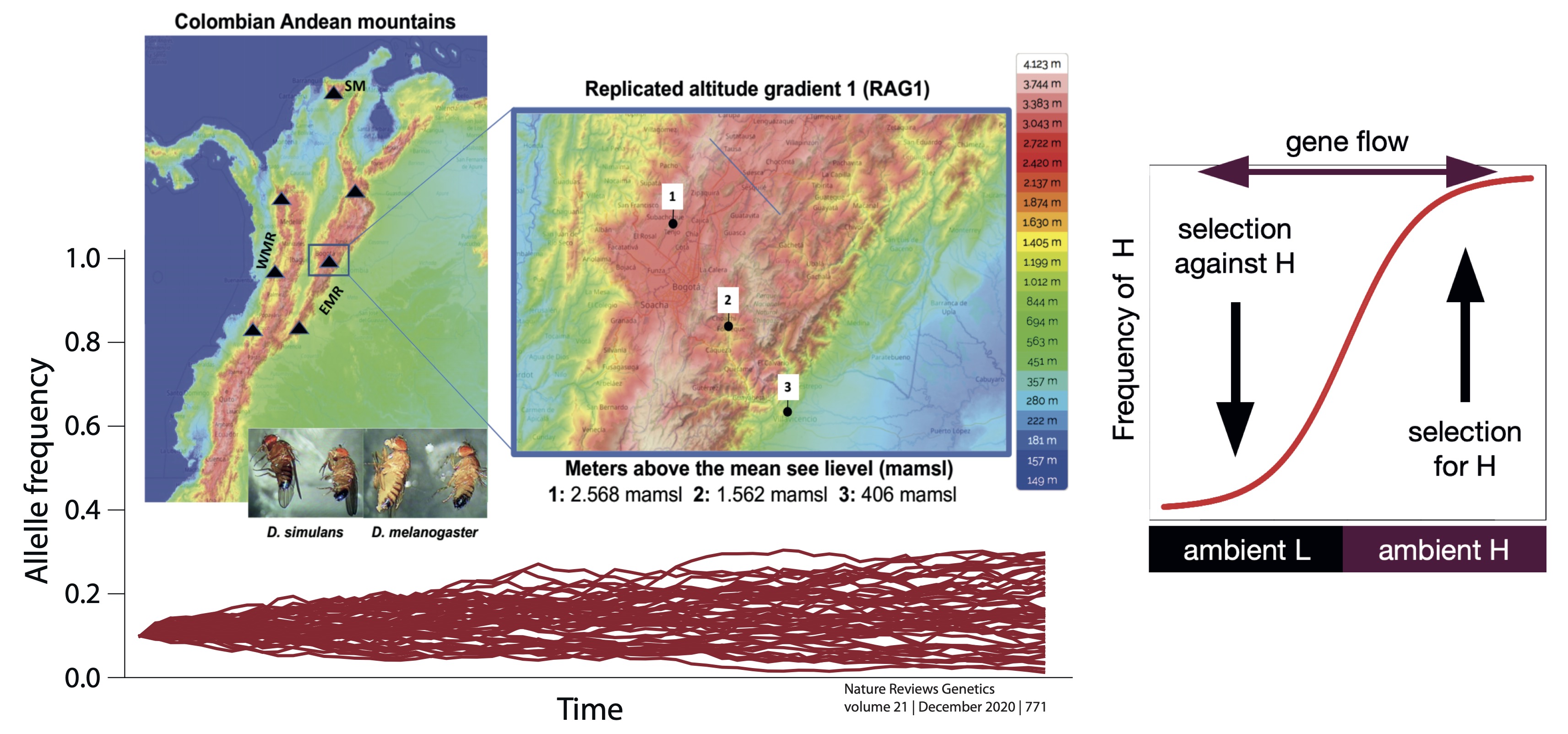
Genomic basis of polygenic adaptation to high-altitude in Drosophila
The main goal of our research group is to understand the molecular mechanisms underlying adaptation in natural populations. Our case study are altitude gradients (i.e., changes in elevation), an ecological limit with important variations in temperature and humidity along very short distances, typically on a few kilometres. Our research model is Drosophila, specifically two species with large effective population sizes and high-quality genome sequences and annotations, two aspects that are determinants to find and characterize the footprint of positive selection at the molecular level. We are combining high-throughput sequencing data, powerful population genomics and bioinformatics inference, and computer modelling, in an innovative large-scale study across more than 2,500km at Colombian Andes Mountain ranges. We are focusing our study on determining the role of polygenic adaptation, i.e., the process in which a population adapts through subtle changes in allele frequencies at many functionally relevant sites across the genome (hundreds or thousands of genes) to altitude in Colombian populations and its environmental drivers.
 Tasks to be carried out by the student:
Tasks to be carried out by the student:
Expected student skills:
Basic knowledge on evolutionary genetics, and/or on NGS data handling and analysis. Experience with Linux operating systems, and one of the programing languages commonly used in bioinformatics (Perl, Python, R) are desirable.Project supervisor:
Alejandro Sánchez-Gracia (elsanchez@ub.edu)See other projects in the EG&B Web page.
Software developed in the research group:
Publications of the EGB research group: