 BSc and MSc Thesis Projects -2022
BSc and MSc Thesis Projects -2022
Oferta de pràcticums, TFG i TFM a la Facultat de Biologia
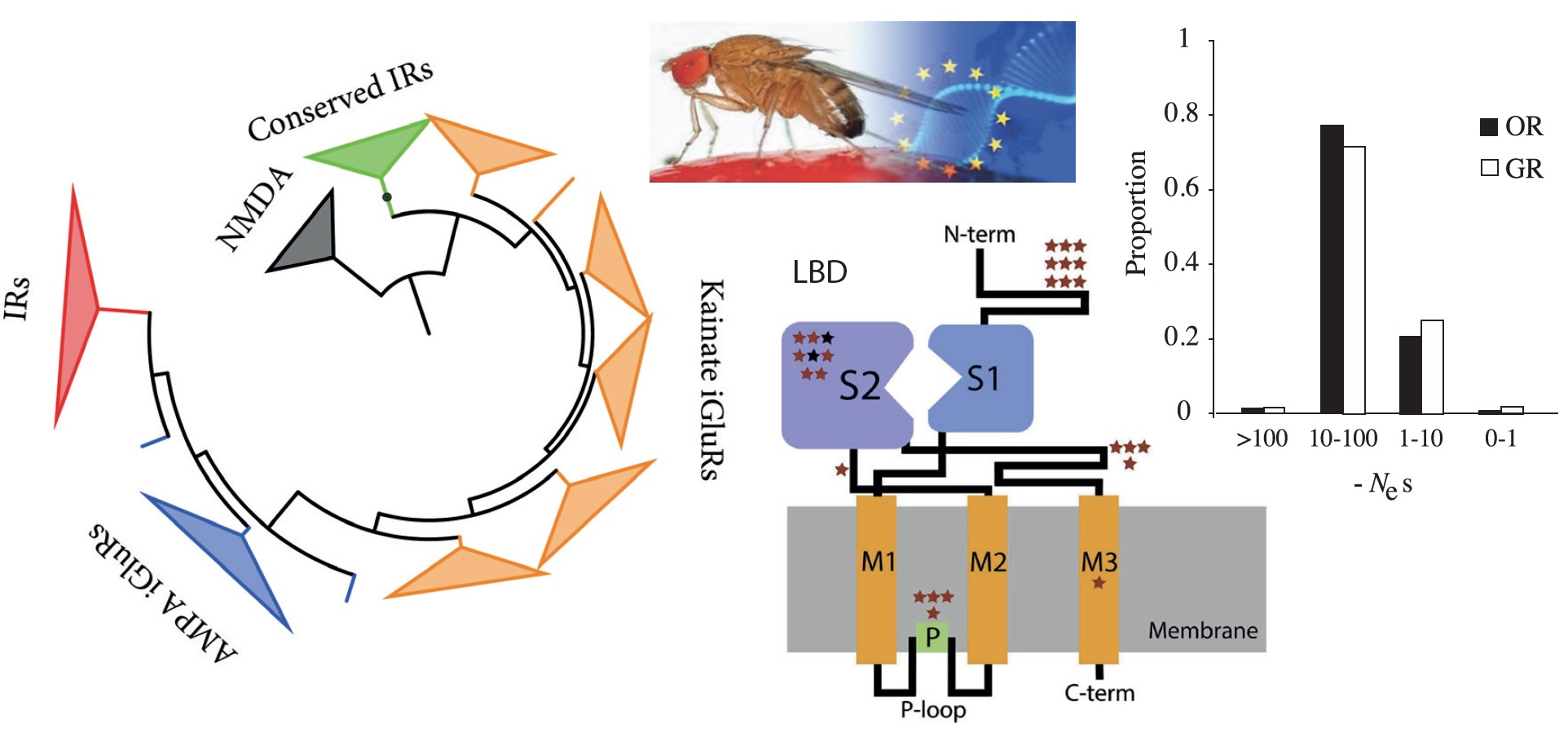
Molecular evolution of animal chemosensory-related gene families
My research group is interested in to explore how functional divergence and pleiotropy affects the organization, structure, and molecular evolution of multigene families in animal genomes, with a special focus on their consequences for adaptation.
My research model are the gene families encoding insect chemoreceptors (i.e., odorant, gustatory and ionotropic receptors).
My approximation to the problem is rooted in comparative and population genomics and I use integrated computational and analytical tools to address evolutionary and mechanistic questions.
As a member of the European Drosophila Population Genomics consortium (DrosEU), we have access to population genomics data from a large set of populations distributed across the continent, including different latitudes (and climates) and seasonal data.
This specific subproject aims to investigate the relationship between the distribution of fitness effects (DFE) of mutations (both deleterious and adaptive) in the three main chemoreceptor families, and pleiotropy (i.e., the number of traits affected by these mutations, which will be approached by gene expression data available for the members of these families).
 Tasks to be carried out by the student:
Tasks to be carried out by the student:
Expected student skills:
Basic knowledge on molecular population genetics, and/or on NGS data handling and analysis. Experience with Linux operating systems, and one of the programing languages commonly used in bioinformatics (Perl, Python, R) are desirable.Project supervisor:
Alejandro Sánchez-Gracia (elsanchez@ub.edu)See other projects in the EG&B Web page.
Software developed in the research group:
Publications of the EGB research group: